First record of Unionicola (Myanmaratax) savadiensis from India, with remarks on parasite-host relationship in unionicolid mites (Acari, Unionicolidae)
Chatterjee, Tapas  1
; Khan, Ajaz Ali Ahmed
1
; Khan, Ajaz Ali Ahmed  2
; Singh, Ravail
2
; Singh, Ravail  3
; Vidrine, Malcolm
3
; Vidrine, Malcolm  4
; Zawal, Andrzej
4
; Zawal, Andrzej  5
and Pešić, Vladimir
5
and Pešić, Vladimir  6
6
1Near Hari Mandir Road, Hirapur, Dhanbad 826001, Jharkhand, India.
2CSIR-Indian Institute of Integrative Medicine, Jammu 180001, India & Academy of Scientific and Innovative Research (ACSIR), Gaziabad- 201002, India.
3✉ CSIR-Indian Institute of Integrative Medicine, Jammu 180001, India & Academy of Scientific and Innovative Research (ACSIR), Gaziabad- 201002, India.
4Division of Arts and Sciences, Louisiana State University Eunice, Eunice, LA 70535, USA.
5Institute of Marine and Environmental Sciences, Center of Molecular Biology and Biotechnology, University of Szczecin, Wąska 13, 71–415 Szczecin, Poland.
6Department of Biology, University of Montenegro, Cetinjski put b.b., 81000 Podgorica, Montenegro.
2023 - Volume: 63 Issue: 4 pages: 1094-1101
https://doi.org/10.24349/lb8h-imelOriginal research
Keywords
Abstract
Introduction
The majority of the 265 known unionicolid freshwater mites (Acari: Unionicolidae: Unionicola) are associated with molluscs during at least one stage of their life cycle. The mites are currently known to infest five of the six families of freshwater mussels (Bivalvia: Unionoida) (Edwards and Vidrine 2013a, b). The mollusc-associated mites are readily divided into two groups: mantle mites (the mantle and foot tissue of their hosts as sites of oviposition) and gill mites (the gill tissues of their hosts as sites of oviposition) (Edwards and Vidrine 2006, 2013a, b, 2020). In the Northern Hemisphere, it is not uncommon to find the representatives of both groups of mites in a single host individual, however, the mites apparently are highly specific as to not only as sites of oviposition but also as to their selection of host species. The water mite literature supporting these hypotheses was summarized in Edwards and Vidrine (2013a, b, 2021), and Ernsting et al. (2014), and more recently in Chapurina et al. (2021, 2022a, b). Discovery of species new to science as well as associations new to science are also not uncommon, however, each discovery is important and helps to qualify significant hypotheses in evolutionary ecology of the complex 200-million year history of mites and molluscs associations (Edwards and Vidrine 2020). The substantiation of this history with molecular evidence from a variety of sources, including both molluscan and acarine DNA sequences, is important.
In historical Indian collections, Karl Viets (1926) described two mites from a single host species in India. Lamellidens marginalis (Lamarck, 1819) was infected by Unionicola (Prasadatax) diversipes Viets, 1926 (a gill mite) and U. (Imamuratax) scutigera Viets, 1926 (a mantle mite). Also, in Majumder's collections (Majumder and Pal 1987, 1988, 1990), U. diversipes, U. scutigera, and Unionicola sp. (= U. (Majumderatax) hankoi Szalay, 1927) (a mantle mite) were found in the same host species. John and Inasu (2004) reported U. diversipes and U. (Myanmaratax) brandti Vidrine, 1985 from the same host species. It is possible that the latter record was actually a mite belonging to one of the species described in the new subgenus in Chapurina et al. (2022a), but lacking evidence supporting this, the record is considered doubtful. Chapurina et al. (2022a, b) reported a number of species from Lamellidens Simpson, 1900 as gill mites from Myanmar, including U. diversipes, U. (Dimockatax) huangthayaensis (Chapurina et al., 2022b), U. (Myanmaratax) generosa Chapurina, Vidrine, Kondakov, Vikhrev & Bolotov, 2022 (in Chapurina et al., 2022a) and U. (Myanmaratax) savadiensis Chapurina, Vidrine, Kondakov, Vikhrev & Bolotov, 2022 (in Chapurina et al., 2022a).
The records of U. (M.) savadiensis in new host species, accompanied with molecular analyses, contribute significantly to our global picture of the co-evolution of mussels and mites. The series of co-phylogenetic hypotheses is discussed elsewhere (Edwards and Vidrine 2013a), and our focus here is on the mites of the genus Lamellidens (Unionidae: Parreysiinae, Lamellidentini).
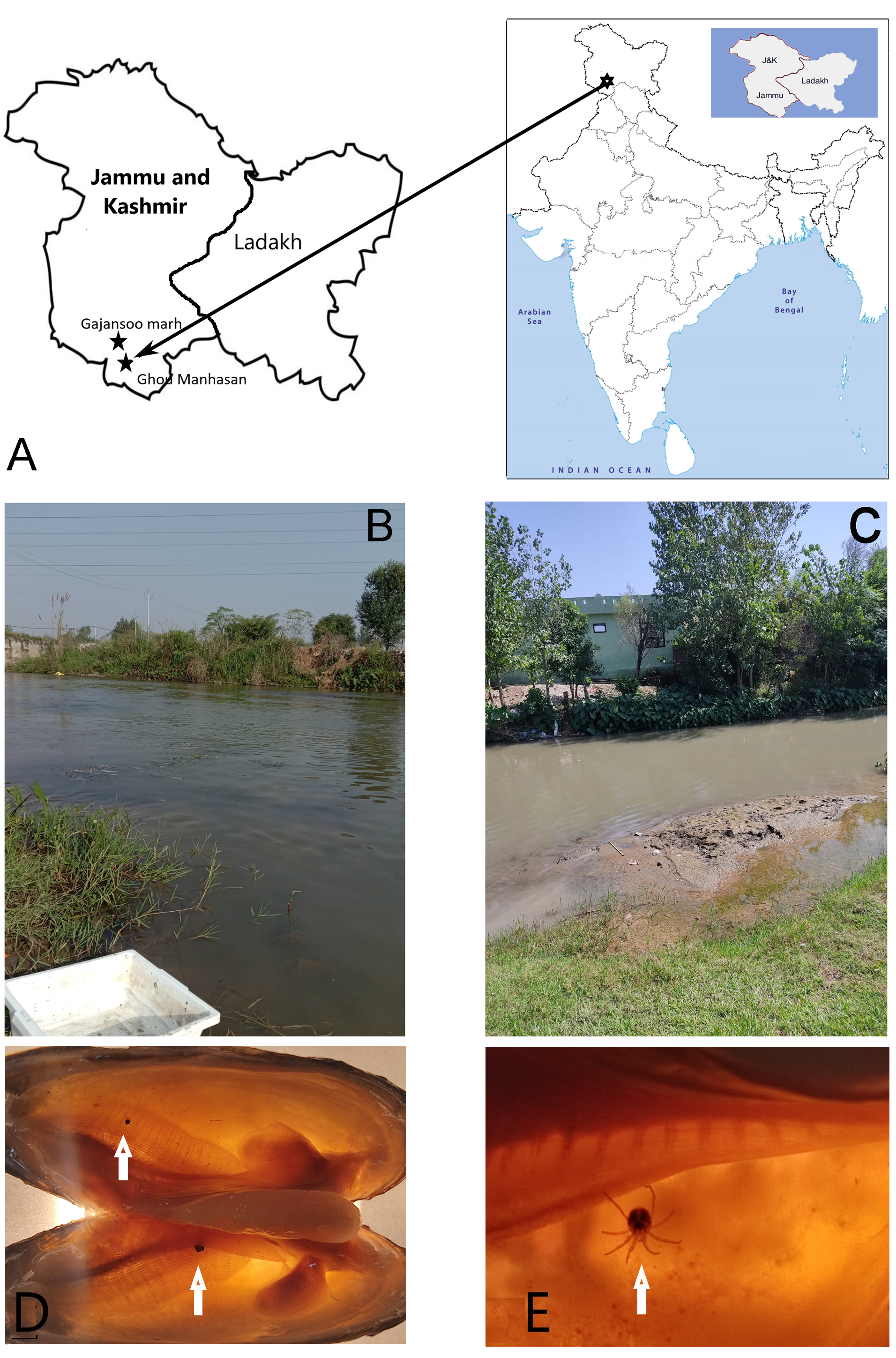
Material and methods
The unionid freshwater mussels were collected by the second author (AAAKK) from two sites in Jammu and Kashmir of India in September-October 2022: 1/. Gou Manhasan, a tributary of River Chenab (32.7232°N; 74.7334 °E), 20 km from Jammu town and Gajansoo Marh, and 2/. a tributary of River Chenab (32.7651°N, 74.7150°E), 12 km from Jammu town - about 2 m from the bank of the river at a depth of 5 cm in the sediment in Jammu and Kashmir of India (Fig. 1B-C). Mites were collected from gills of Corbicula cashmiriensis Deshayes, 1854, Lamellidens corrianus (Lea, 1834), and L. marginalis by manual dissection in a closed chamber using autoclaved surgical blade, forceps and needles (Fig. 1D-E). Mites were taken from the upper layer of gills of C. cashmiriensis and L. marginalis, and from a deeper layer of gills of L. corianus. The mite samples were preserved in 96% ethanol immediately after collecting. For morphological research, permanent slides of mite specimens were prepared.


After DNA extraction, the specimen vouchers were morphologically examined. Some of these vouchers were dissected and slide mounted in Faure's medium, while the rest was transferred to Koenike-fluid and stored in the private collection of Vladimir Pešić. DNA sequences prepared in the course of this study were deposited in BOLD (The Barcode of Life Data System) and GenBank (see Tab. 1). The DNA extracts were archived at −80 °C at the Centre for Biodiversity Genomics in Guelph, Canada (CBG; biodiversitygenomics.net).
Molecular analyses were conducted at the Canadian Centre for DNA Barcoding, Guelph, Ontario, Canada (CCDB; http://ccdb.ca/ ![]() ). The specimens were sequenced for the barcode region of COI using standard invertebrate DNA extraction (Ivanova et al. 2007), amplification (Ivanova and Grainger 2007a) and sequencing protocols (Ivanova and Grainger 2007b).
). The specimens were sequenced for the barcode region of COI using standard invertebrate DNA extraction (Ivanova et al. 2007), amplification (Ivanova and Grainger 2007a) and sequencing protocols (Ivanova and Grainger 2007b).
The final dataset included 28 COI sequences of the genus Unionicola: U. (M.) savadiensis (n=8), U. generosa (n=4), U. trapezidens (n=4), U. robacki (n=9), U. sella (n=2) and U. ypsilophora (n=1). COI sequence data were additionally taken from Chapurina et al. (2021, 2022a) and Pešić et al. (2021), and downloaded from the respective sequence data archives. Sequences were aligned with MUSCLE (Edgar 2004). MEGA X software (Kumar et al. 2018) was used to calculate Neighbor-Joining NJ trees based on the Kimura 2-parameter (K2P; Kimura 1980) distances (standard for barcoding studies) and pair wise deletion of missing data. The support for tree branches was calculated by the nonparametric bootstrap method (Felsenstein 1985) with 1000 replicates and shown next to the branches. Codon positions included were 1st+2nd+3rd+Noncoding. All ambiguous positions were removed for each sequence pair.
Results
Molecular Analysis
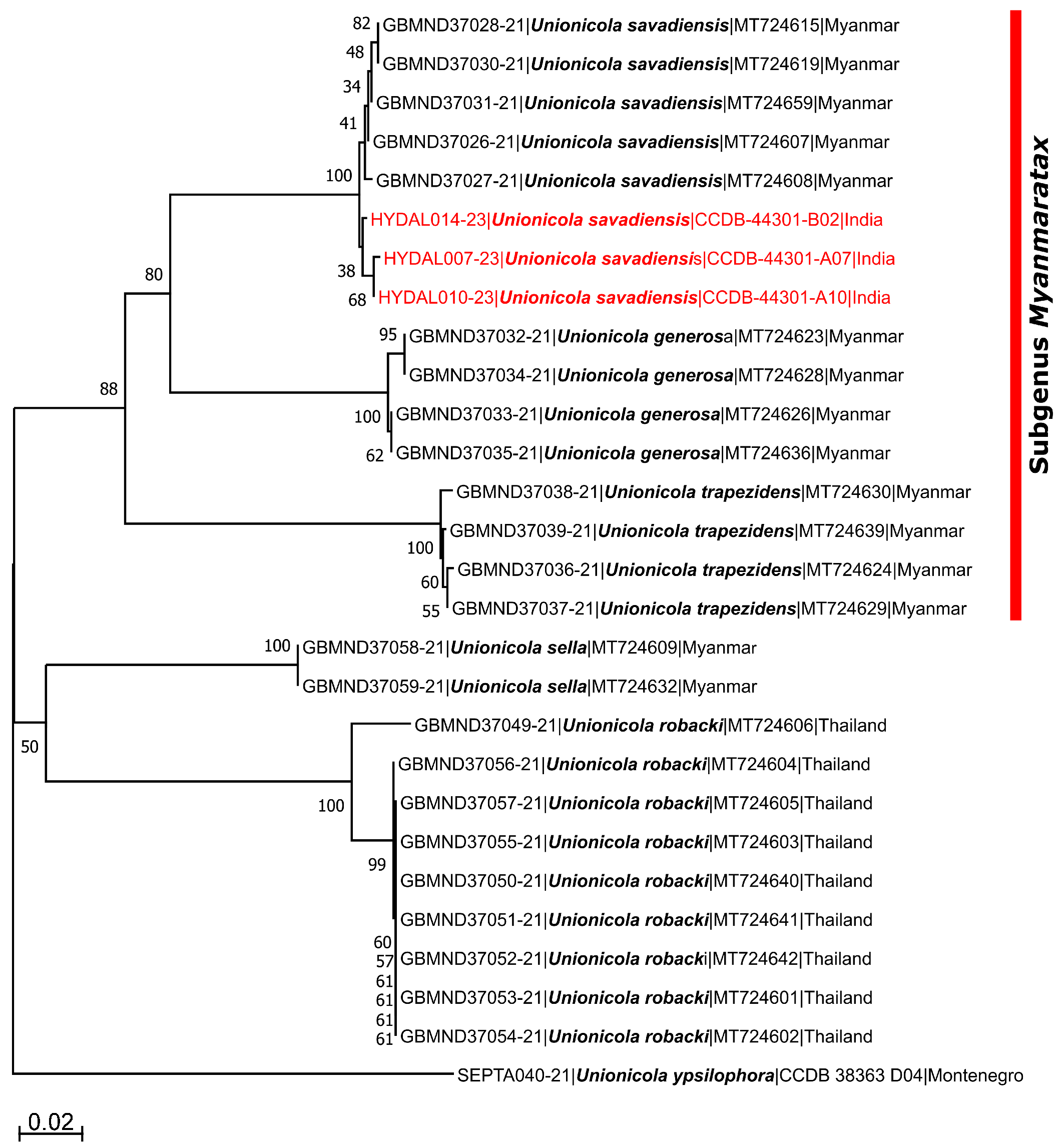
A 639-658 bp fragments for three specimens of Unionicola collected from mussels were successfully amplified. Species identification of studied specimens based on BOLD and GenBank databases is shown in Table 1. Three specimens were identified to species level as they showed sequence matches of 99.53-99.54% with U. (M.) savadiensis sequences available in GenBank database. For another three specimens, morphologically attributed to U. savadiensis (two specimens collected in the gills of C. cashmiriensis from Gou Manhasan and one specimen collected in the gills of L. marginalis from Gajansoo Marh; Tab. 1), we amplified and sequenced DNA of the mussel L. corrianus instead of water mite DNA. These sequences showed sequence match of 99.65-99-67% with L. corrianus (AF339457) from India submitted by Lopes-Lima et al. (2021).
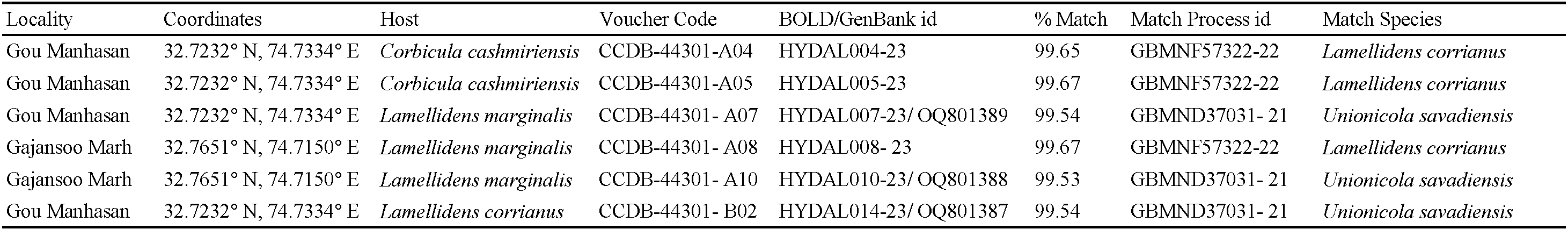


To further confirm the taxonomic status of the examined specimens, 28 COI Unionicola sequences were downloaded from the BOLD database. The Neighbor Joining (NJ) tree was constructed in MEGA X software and displayed in Figure 2. The sequences retrieved from Unionicola specimens from Jammu and Kashmir, formed a distinct cluster with sequences of U. (M.) savadiensis from Myanmar (Fig. 2).


Systematics
Family Unionicolidae Oudemans, 1909
Genus Unionicola Haldeman, 1842
Subgenus Myanmaratax Chapurina, Vidrine, Kondakov, Vikhrev & Bolotov, 2022
Unionicola (Myanmaratax) savadiensis Chapurina, Vidrine, Kondakov, Vikhrev & Bolotov, 2022
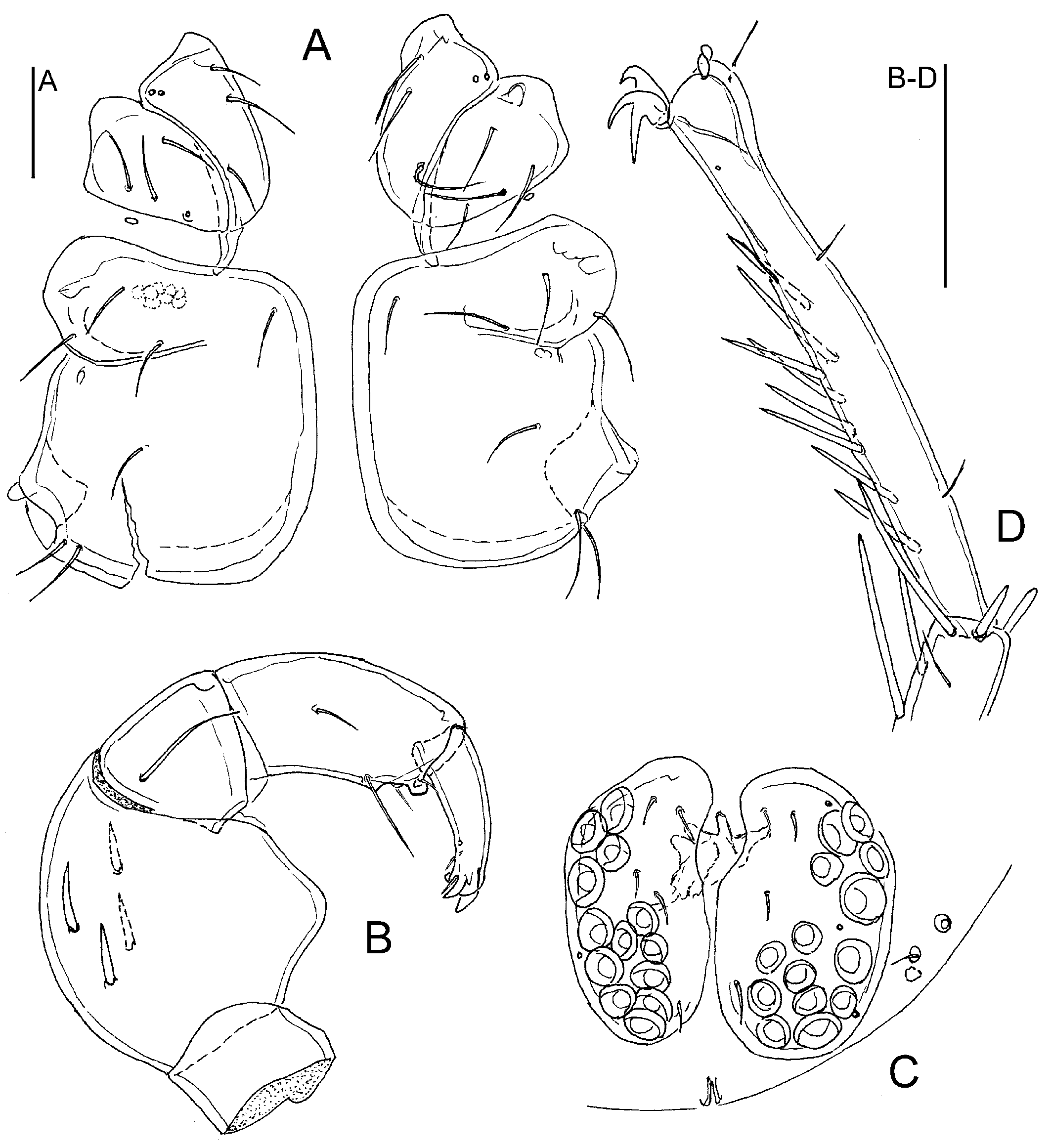
Figure 3A-D
Material examined — Jammu and Kashmir: CC Vial 1, Gou Manhasan, a tributary of River Chenab, 32.7232°N, 74.7334 °E, from the gills of C. cashmiriensis, 2♀; CC Vial 2, ibid., from the gills of C. cashmiriensis, 1♂, dissected and slide mounted (RMNH); CC Vial 3, ibid., from the gills of C. cashmiriensis, 2♀; LMM Vial 1, Gou Manhasan, a tributary of River Chenab, 32.7232°N, 74.7334°E, from the gills of L. marginalis, 2♂ (1♂ sequenced, Tab. 1); LMM Vial 2, Gajansoo Marh, a tributary of River Chenab, 32.7651°N, 74.7150°E, from the gills of L. marginalis, 4♂, 1♀ (1♂ sequenced, Tab. 1), 1♂ dissected and slide mounted (RMNH); LCM Vial 1: Gajansoo Marh, a tributary of River Chenab, 32.7651°N, 74.7150°E, from the gills of L. corrianus, 1♂, 3♀; LCM Vial 2, Gou Manhasan, a tributary of River Chenab, 32.7232°N, 74.7334°E, from the gills of L. corrianus, 2♂, 2♀ (1♀ sequenced, Tab. 1).
Remarks — Unionicola (M.) savadiensis was originally described from the Indaw Lake in Myanmar. The holotype was collected from the gills of the freshwater mussel, L. savadiensis (Nevill, 1877), and the paratypes from the gills of the type host species and of L. generosus (Gould, 1847). Chapurina et al. (2022a) assigned U. savadiensis to the newly described subgenus Myanmaratax Chapurina et al. 2022, which, in addition to U. (M.) savadiensis, includes three more mussel-associated gill mites, i.e., U. (M.) brandti Vidrine, 1985, U. (M.) generosa, and U. (M.) trapezidens Chapurina, Vidrine, Kondakov, Vikhrev & Bolotov, 2022. Members of this subgenus use freshwater mussel (Bivalvia: Unionidae) species of the genera Lamellidens and Trapezidens in Western Indochina as well as Contradens and Ensidens in the Sundal and Subregion (Chapurina et al. 2022a) as the primary hosts.
The collected specimens from Jammu and Kashmir match the original description of U. (M.) savadiensis in regard to shapes of coxal plates and genital fields (Fig. 3A, C), palps (Fig. 3B) and terminal segments of the fourth legs (Fig. 3D).


Discussion
In this study, U (M.) savadiensis was found to be associated with three species of molluscs: L. marginalis, L. corrianus and Corbicula cashmiriensis. Our data reveal a much wider range extension of U. (M.) savadiensis and its mussel host, and fit the hypotheses of coevolution and the rather tight host-parasite association within Asian mussels, a phenomenon that has also been proven for gill mites and their unionid mussel hosts in North America (Edwards and Vidrine 2006, 2013b, 2020; Edwards et al. 2010; Ernsting et al. 2014).
We tend to refute the discovery of the genus Corbicula serving as a host for U. (M.) savadiensis. Unionicola mites have not been verified from the family Cyrenidae Gray, 1840, which includes the genus Corbicula. Spurious records however exist, including records from Egypt (Abdel-Garber et al. 2018)—these were apparently compromised as mussels, including Corbicula and infected unionoid mussels, were held in the same container for transport and probably resulted in vagrancy among these mites. Vagrancy is common when mussels of different species hosting different mites are held together under stress (Edwards and Vidrine 2013a). We suspect that this also accounts for our current observation in India. We showed that all DNA sequences of mussels, which were amplified instead of mite DNA, belong to L. corrianus, also from Gou Manhasan (where C. cashmiriensis is listed as a ''host'') and from the Gajansoo Marh (where L. marginalis is listed as a ''host''). Therefore, we can speculate that the members of U. savadiensis in all three localities parasitize L. corrianus, but occasionally can occur also on two other species (without feeding on them), and their presence on the latter species should be treated as vagrant associations. This is confirmed by the observation that mites were found on the surface of gill in L. marginalis and C. cashmiriensis, while mites were found in deeper layers of the gill in L. corrianus.
Secondly, in addition to discussion of coevolution, the adaptations to new hosts, i.e.'host-switching' (Cichy et al. 2016), must be taken into account. Such a situation may occur in the case of Unionicola ypsilophora (Bonz, 1783), which inhabits invasive Sinanodonta woodiana (Lea, 1834) in Poland (Cichy et al. 2016). Unlike'vagrancy' — where mites move from one species of mussel to another species,'host-switching' (also known as'Association by Colonization' in studies of coevolution and cophylogeny) is an uncommon phenomenon, at least in the literature. However, some mite interactions can only be explained by incidences of'host-switching,' with the best examples being the occurrences of unionicolid mites in freshwater snails. These snail mites are apparently derived from mussel mites, and this is evident in the morphological and molecular studies by Wu et al. (2009, 2012) in Chinese mussel and snail mites (see also Edwards and Vidrine 2013a). In this study, we do not suggest that'host-switching' has or may be occurring among our Indian findings, but we do want to note that it was considered.
Acknowledgement
The second and third authors (Ajaz Ali Ahmed Khan and Ravail Singh) want to thank the Director of Indian Institute of Integrative Medicine, Jammu for providing facilities to perform their work (publication database number is CSIR-IIIM/IPR/00585). The last author (Vladimir Pešić) wishes to thank Milica Jovanović and Ana Manović (University of Podgorica) for their excellent help with barcoding examined mites.
References
- Abdel-Gaber R., Fol M., Quraishy A. 2018. Light and scanning electron microscopic studies of Unionicola tetrafurcatus (Acari: Unioncolidae) infecting four freshwater bivalve species with referring to histopathological effect on its hosts. J. Parasitol., 104: 359-371. https://doi.org/10.1645/18-31
- Chapurina Y.E., Bolotov I.N., Vidrine M.F., Vikhrev I.V., Lunn Z., Chan N., Win T., Bespalaya Y.V., Aksenova O.V., Gofarov M.Y., Kondakov A.V., Konopleva E.S. 2021. Taxonomic richness and host range of tropical Asian mussel-associated mite assemblage (Acari: Unionicolidae) with description of a new subgenus and species of parasitic mite from freshwater pearl mussels (Unionida: Margaritiferidae). J. Zool. Syst. Evol. Res., 59: 613-634. https://doi.org/10.1111/jzs.12445
- Chapurina Y.E., Konopleva E.S., Vidrine M.F., Vikhrev I.V., Lunn Z., Chan N., Win T., Kondakov A.V., Zubrii N.A., Bespalaya Y.V., Aksenova O.V., Gofarov M.Y., Bolotov I.N. 2022a. New molecular-based phylogeny of mussel-associated mites reveals a new subgenus and three new species representing an example of a host-driven radiation in Indochina and confirms the concept of division of the genus Unionicola Haldeman, 1842 (Acari: Unionicolidae) into numerous subgenera. Diversity, 14: 848. https://doi.org/10.3390/d14100848
- Chapurina Y.E., Kondakov A., Chan N., Vikhrev I.V., Bolotov I.N., Konopleva E.S., Win T., Lunn Z.A. 2022b. New species Unionicola (Dimockatax stat. rev.) haungthayawensis sp. nov. (Trombidiformes: Unionicolidae) from the freshwater mussel Lamellidens generosus (Gould, 1847) in Myanmar. Ecologica Montenegrina, 56: 28-39. https://doi.org/10.37828/em.2022.56.4
- Cichy A., Urbanska M., Marszewska A., Andrzejewski W., Zbikowska E. 2016. The invasive Chinese pond mussel Sinanodonta woodiana (Lea, 1834) as a host for native symbionts in European waters. J. Limnol., 75: 288-296. https://doi.org/10.4081/jlimnol.2016.1334
- Edgar R.C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high 679 throughput. Nucleic Acids Res., 32(5): 1792-1797. https://doi.org/10.1093/nar/gkh340
- Edwards D.D., Vidrine M.F. 2006. Host specificity among Unionicola spp. (Acari: Unionicolidae) parasitizing freshwater mussels. J. Parasitol., 92: 977-983. https://doi.org/10.1645/GE-3565.1
- Edwards D.D., Vidrine M.F. 2013a. Mites of freshwater mollusks. M. F. Vidrine (Eunice, LA). 332 pp.
- Edwards D.D., Vidrine M.F. 2013b. Patterns of species richness among assemblages of Unionicola spp. (Acari: Unionicolidae) inhabiting freshwater mussels (Bivalvia: Unionoida) of North America. J. Parasitol., 99: 212-217. https://doi.org/10.1645/GE-3208.1
- Edwards D. D., Vidrine M.F. 2020. Host diversity affects parasite diversity: A case study involving Unionicola spp. inhabiting freshwater mussels. J. Parasitol.,106: 675-678. https://doi.org/10.1645/19-190
- Edwards D.D., Ernsting B.R., Vidrine M.F. 2010. Phylogenetic relationships among Unionicola (Acari: Unionicolidae) mussel-mites of North America based on mitochondrial cytochrome oxidase I sequences. Zootaxa, 2537: 47-57. https://doi.org/10.11646/zootaxa.2537.1.4
- Ernsting B.R., Edwards D.D., Timbrook T.A., Frerichs M.M. 2014. Preliminary evidence of cryptic species among host-associated populations of Unionicola hoesei (Acari: Unionicolidae). Internat. J. Acarol., 40: 358-365. https://doi.org/10.1080/01647954.2014.919359
- Felsenstein J.1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 39: 783-791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
- Ivanova N.V., Grainger C.M. 2007a. CCDB protocols, COI amplification. Available from: https://ccdb.ca/site/wp-content/uploads/2016/09/CCDB_Amplification.pdf (Accessed 20 Dec. 2021)
- Ivanova N.V., Grainger C.M. 2007b. CCDB protocols, sequencing. Available from: https://ccdb.ca/site/wp-content/uploads/2016/09/CCDB_Sequencing.pdf (Accessed 20 Dec. 2021)
- Ivanova N.V., de Waard J.R., Hebert P.D.N. 2007. CCDB protocols, glass fiber plate DNA extraction. Available from: https://ccdb.ca/site/wp-content/uploads/2016/09/CCDB_DNA_Extraction.pdf (Accessed 20 Dec.)
- John C.C., Inasu N.D. 2004. Water mites associated with the fresh water mussel Lamellidens marginalis (Pelecypoda: Unionidae). Zoos' Print J., 20: 1986-1987. https://doi.org/10.11609/JoTT.ZPJ.1135.1986-7
- Kimura M.1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol., 16: 111-120. https://doi.org/10.1007/BF01731581
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol., 35: 1547-1549. https://doi.org/10.1093/molbev/msy096
- Lopes-Lima M., Gürlek M.E., Kebapçı Ü., Şereflişan H., Yanık T., Mirzajani A., Neubert E., Prié V., Teixeira A., Gomes-Dos-Santos A., Barros-García D., Bolotov I.N., Kondakov A.V., Vikhrev I.V., Tomilova A.A., Özcan T., Altun A., Gonçalves D.V., Bogan A.E., Froufe E. 2021. Diversity, biogeography, evolutionary relationships, and conservation of Eastern Mediterranean freshwater mussels (Bivalvia: Unionidae). Mol. Phylogenet. Evol., 163: 107261. https://doi.org/10.1016/j.ympev.2021.107261
- Majumder M.Z.R., Pal S.G. 1987. Larval development of a Unionicola sp., a freshwater mite of a freshwater bivalve (Lamellidens marginalis) from Bengal. In: Palanichamy S. (ed.) Proceedings Fifth Indian Symp. Invert. Reprod., 158-170.
- Majumder M.Z.R., Pal S.G. 1988. Adaptations of Unionicola sp. A freshwater mite on Lamellidens marginalis from Bengal. Bicovas, 1: 191-202.
- Majumder M.Z.R., Pal S.G.1990. Egg laying behaviour of Unionicola sp. in freshwater bivalve host. Proc. Nat. Symp. Anim. Behav., In: Patel B.H. (ed.), The Behaviour Sir P. P. Institute of Science, Bhavnagar, 1989, 83-88.
- Pešić V., Zawal A., Manović A., Bańkowska A., Jovanović M. 2021. A DNA barcode library for the water mites of Montenegro. Biodivers. Data J., 9: e78311. https://doi.org/10.3897/BDJ.9.e78311
- Viets K. 1926. Indische Wassermilben. Zool. Jb. Syst., 52: 371-394.
- Wu H.B., Hu B., Zhu Z., Wen C. 2009. Cladistic analysis on the genus Unionicola (Acari: Unionicolidae) from China. Entomotaxonomia, 31: 152-160.
- Wu H.B., Wen C.G., Guo W. 2012. Sequence variation of the mitochondrial 12S rRNA gene among Unionicola (Wolcottatax) arcuata (Acari: Unionicolidae) from freshwater mussels in China. Internat. J. Acarol., 38: 394-401. https://doi.org/10.1080/01647954.2012.657241



2023-05-04
Date accepted:
2023-09-26
Date published:
2023-10-10
Edited by:
Mąkol, Joanna

This work is licensed under a Creative Commons Attribution 4.0 International License
2023 Chatterjee, Tapas; Khan, Ajaz Ali Ahmed; Singh, Ravail; Vidrine, Malcolm; Zawal, Andrzej and Pešić, Vladimir
Download the citation
RIS with abstract
(Zotero, Endnote, Reference Manager, ProCite, RefWorks, Mendeley)
RIS without abstract
BIB
(Zotero, BibTeX)
TXT
(PubMed, Txt)



